Abstract
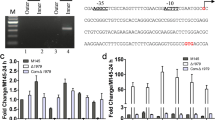
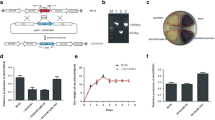
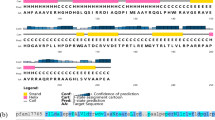
Many regulatory genes that affect cellular development in Streptomyces, such as the canonical bld genes, have already been identified. However, in this study, we identified sven_5003 in Streptomyces venezuelae as a major new developmental regulatory gene, the deletion of which leads to a bald phenotype, typical of bld mutants, under multiple growth conditions. Our data indicated that disruption of sven_5003 also has a differential impact on the production of the two antibiotics jadomycin and chloramphenicol. Enhanced production of jadomycin but reduced production of chloramphenicol were detected in our sven_5003 mutant strain (S. venezuelae D5003). RNA-Seq analysis indicated that SVEN_5003 impacts expression of hundreds of genes, including genes involved in development, primary and secondary metabolism, and genes of unknown function, a finding confirmed by real-time PCR analysis. Transcriptional analysis indicated that sven_5003 is an auto-regulatory gene, repressing its own expression. Despite the evidence indicating that SVEN_5003 is a regulatory factor, a putative DNA-binding domain was not predicted from its primary amino acid sequence, implying an unknown regulatory mechanism by SVEN_5003. Our findings revealed that SVEN_5003 is a pleiotropic regulator with a critical role in morphological development in S. venezuelae.





Similar content being viewed by others
References
Hopwood DA (ed) (2007) Streptomyces in nature and medicine. Oxford University Press, Oxford
Chater K (2011) Streptomyces: molecular biology and biotechnology. Caister Academic Press, Poole, pp 43–86
Elliot MA, Buttner MJ, Nodwell, J.R. (2008) In: Whiteworth DE (ed) Myxobacteria: Multicellularity and Differentiation American Society for Microbiology. p 419–438.
Chater KF (2001) Regulation of sporulation in Streptomyces coelicolor A3(2): a checkpoint multiplex? Curr Opin Microbiol 4:667–673
Chandra G, Chater KF (2008) Evolutionary flux of potentially bldA-dependent genes containing the rare leucine codon TTA. Anton Leeuw Int J G 94:111–126
Pope MK, Green B, Westpheling J (1998) The bldB gene encodes a small protein required for morphogenesis, antibiotic production, and catabolite control in Streptomyces coelicolor. J Bacteriol 180:1556–1562
Hunt AC, Servín-González L, Kelemen GH, Buttner MJ (2005) The bldC developmental locus of Streptomyces coelicolor encodes a member of a family of small DNA-binding proteins related to the DNA-binding domains of the MerR family. J Bacteriol 187:716–728
Elliot M, Damji F, Passantino R, Chater K, Leskiw B (1998) The bldD gene of Streptomyces coelicolor A3(2): a regulatory gene involved in morphogenesis and antibiotic production. J Bacteriol 180:1549–1555
Elliot MA, Bibb MJ, Buttner MJ, Leskiw BK (2001) BldD is a direct regulator of key developmental genes in Streptomyces coelicolor A3(2). Mol Microbiol 40:257–269
Elliot MA, Leskiw BK (1999) The BldD protein from Streptomyces coelicolor is a DNA-binding protein. J Bacteriol 181:6832–6835
Tschowri N, Schumacher MA, Schlimpert S, Chinnam NB, Findlay KC, Brennan RG, Buttner MJ (2014) Tetrameric c-di-GMP mediates effective transcription factor dimerization to control Streptomyces development. Cell 158:1136–1147
Bignell DRD, Warawa JL, Strap JL, Chater KF, Leskiw BK (2000) Study of the bldG locus suggests that an anti-anti-sigma factor and an anti-sigma factor may be involved in antibiotic production and sporulation. Microbiol-Sgm 146:2161–2173
Ohnishi Y, Yamazaki H, Kato JY, Tomono A, Horinouchi S (2005) AdpA, a central transcriptional regulator in the A-factor regulatory cascade that leads to morphological development and secondary metabolism in Streptomyces griseus. Biosci Biotechnol Biochem 69:431–439
Nodwell JR, Yang R, Kuo D, Losick R (1999) Extracellular complementation and the identification of additional genes involved in aerial mycelium formation in Streptomyces coelicolor. Genetics 151:569–584
Molle V, Buttner MJ (2000) Different alleles of the response regulator gene bldM arrest Streptomyces coelicolor development at distinct stages. Mol Microbiol 36:1265–1278
Bibb MJ, Molle V, Buttner MJ (2000) Sigma(BldN), an extracytoplasmic function RNA polymerase sigma factor required for aerial mycelium formation in Streptomyces coelicolor A3(2). J Bacteriol 182(16):4606–4616
Al-Bassam MM, Bibb MJ, Bush MJ, Chandra G, Buttner MJ (2014) Response regulator heterodimer formation controls a key stage in Streptomyces development. PLoS Genet 10:e1004554
Zhu Y, Wang X, Zhang J, Ni X, Zhang X, Tao M, Pang X (2022) The regulatory gene wblA is a target of the orphan response regulator OrrA in Streptomyces coelicolor. Environ Microbiol 24:3081–3096
Zhang P, Wu L, Zhu Y, Liu M, Wang Y, Cao G, Chen XL, Tao M, Pang X (2017) Deletion of MtrA inhibits cellular development of Streptomyces coelicolor and alters expression of developmental regulatory genes. Front Microbiol 8:2013
Bentley SD, Chater KF, Cerdeno-Tarraga AM, Challis GL, Thomson NR, James KD, Harris DE, Quail MA, Kieser H, Harper D et al (2002) Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141–147
Chandra G, Chater KF (2013) Developmental biology of Streptomyces from the perspective of 100 actinobacterial genome sequences. FEMS Microbiol Rev 38:345–379
Bush MJ, Bibb MJ, Chandra G, Findlay KC, Buttner MJ (2013) Genes required for aerial growth, cell division, and chromosome segregation are targets of WhiA before sporulation in Streptomyces venezuelae. MBio 4:e00684-e613
Ou X, Zhang B, Zhang L, Zhao G, Ding X (2009) Characterization of rrdA, a TetR family protein gene involved in the regulation of secondary metabolism in Streptomyces coelicolor. Appl Environ Microbiol 75:2158–2165
Kieser T, Bibb MJ, Buttner MJ, Chater KF, Hopwood DA (eds) (2000) Practical streptomyces genetics, 2nd edn. John Innes Foundation, Norwich
Doull JL, Singh AK, Hoare M, Ayer SW (1994) Conditions for the production of jadomycin-B by Streptomyces-Venezuelae Isp5230—effects of heat-shock, ethanol treatment and phage infection. J Ind Microbiol 13:120–125
He Y, Wang Z, Bai L, Liang J, Zhou X, Deng Z (2010) Two pHZ1358-derivative vectors for efficient gene knockout in streptomyces. J Microbiol Biotechnol 20:678–682
Gregory M, Till R, Smith M (2003) Integration site for Streptomyces phage phiBT1 and development of site-specific integrating vectors. J Bacteriol 185:5320–5323
Som NF, Heine D, Holmes NA, Munnoch JT, Chandra G, Seipke RF, Hoskisson PA, Wilkinson B, Hutchings MI (2017) The conserved actinobacterial two-component system MtrAB coordinates chloramphenicol production with sporulation in Streptomyces venezuelae NRRL B-65442. Front Microbiol 8:1145
Zhu Y, Zhang P, Zhang J, Wang J, Lu Y, Pang X (2020) Impact on multiple antibiotic pathways reveals MtrA as a master regulator of antibiotic production in Streptomyces spp. and potentially in other Actinobacteria. Appl Environ Microbiol 86:e01201-01220
Zhu Y, Zhang P, Zhang J, Xu W, Wang X, Wu L, Sheng D, Ma W, Cao G, Chen XL et al (2019) The developmental regulator MtrA binds GlnR boxes and represses nitrogen metabolism genes in Streptomyces coelicolor. Mol Microbiol 112:29–46
Liu M, Zhang P, Zhu Y, Lu T, Wang Y, Cao G, Shi M, Chen XL, Tao M, Pang X (2019) Novel two-component system MacRS is a pleiotropic regulator that controls multiple morphogenic membrane protein genes in Streptomyces coelicolor. Appl Environ Microbiol 85:e02178-e2118
Lu T, Wu X, Cao Q, **a Y, Xun L, Liu H (2022) Sulfane sulfur posttranslationally modifies the global regulator AdpA to influence actinorhodin production and morphological differentiation of Streptomyces coelicolor. Mbio. https://doi.org/10.1128/mbio.03862-21
Lu T, Zhu Y, Zhang P, Sheng D, Cao G, Pang X (2018) SCO5351 is a pleiotropic factor that impacts secondary metabolism and morphological development in Streptomyces coelicolor. FEMS Microbiol Lett. https://doi.org/10.1093/femsle/fny150
Jones SE, Ho L, Rees CA, Hill JE, Nodwell JR, Elliot MA (2017) Streptomyces exploration is triggered by fungal interactions and volatile signals. Elife. https://doi.org/10.7554/eLife.21738
Bush MJ, Chandra G, Findlay KC, Buttner MJ (2017) Multi-layered inhibition of development: BldO is a dedicated repressor of whiB. Mol Microbiol 104:700–711
Claessen D, Stokroos I, Deelstra HJ, Penninga NA, Bormann C, Salas JA, Dijkhuizen L, Wosten HA (2004) The formation of the rodlet layer of streptomycetes is the result of the interplay between rodlins and chaplins. Mol Microbiol 53:433–443
Kodani S, Hudson ME, Durrant MC, Buttner MJ, Nodwell JR, Willey JM (2004) The SapB morphogen is a lantibiotic-like peptide derived from the product of the developmental gene ramS in Streptomyces coelicolor. Proc Natl Acad Sci U S A 101:11448–11453
Tiffert Y, Supra P, Wurm R, Wohlleben W, Wagner R, Reuther J (2008) The Streptomyces coelicolor GlnR regulon: identification of new GlnR targets and evidence for a central role of GlnR in nitrogen metabolism in actinomycetes. Mol Microbiol 67:861–880
Sola-Landa A, Rodriguez-Garcia A, Apel AK, Martin JF (2008) Target genes and structure of the direct repeats in the DNA-binding sequences of the response regulator PhoP in Streptomyces coelicolor. Nucleic Acids Res 36:1358–1368
Funding
This work was supported by grants from the National Natural Science Foundation of China (NSF32270072 to XP) and from the Qingdao Natural Science Foundation (23-2-1-27-zyyd-jch to YZ).
Author information
Authors and Affiliations
Contributions
XP conceived and supervised the study. YZ and TL performed the experiments and collected data. XP, YZ, HZ, and ML analyzed the data. XP wrote the manuscript. All authors read and approved the manuscript.